Research on the development of new drugs generally starts with target selection followed by hit identification, hit-to-lead confirmation, lead optimization and clinical candidate selection. Virtual screening has been widely applied in early-stage drug discovery. As an alternative or complementary approach to high-throughput screening (HTS) assays with high cost and low hit rate, virtual screening is an efficient computational method to identify drug candidates in silico from large chemical compound databases. Its usefulness has been verified by current applications that successfully retrieved hit and lead identifications against various disease targets.
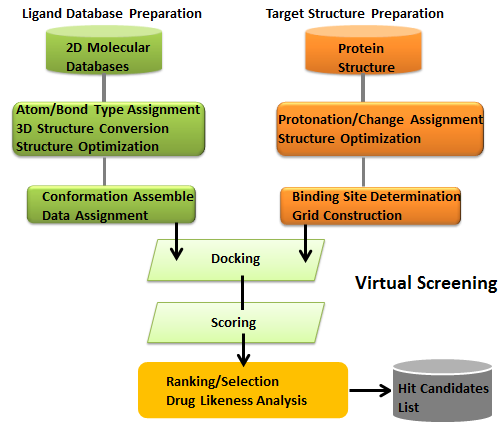
Virtual screening is a knowledge-driven and detailed compound database searching approach which attempts to find the novel compounds and chemotypes with a required biological activity as alternatives to existing ligands, or sometimes to make first inroads into finding ligands for unexplored putative drug targets for which crystal structures, solution structures or high confidence homology models are available. When the three-dimensional (3D) structure of a target is unknown, ligand-based virtual screening can be applied as a compound selection filter for the identification of biologically active compounds. Ligand-based virtual screening involves two different methods: i) flexible alignment of molecules by considering only the atomic contributions; and ii) use of other chemical features that are unrelated to 3D pharmacophore representations, such as hydrogen bonding and lipophilicity, as the input data for flexible alignment. In contrast, if the 3D structure of a target protein is available, both high-throughput docking and receptor-based pharmacophore virtual screening can be applied for identifying new drug candidates.
Virtual screening can be divided into two broad categories, namely structure-based virtual screening (SBVS) and ligand-based virtual screening (LBVS).
BOC Sciences can provide you high-quality service using the virtual screening method in early-stage drug discovery. According to your requirements, we find out compounds with a required biological activity in the database. With the virtual screening method, virtually millions of compounds can be screened for hit identification, which can improve your efficiency of drug discovery.
Our virtual screening integrates computational models and large chemical databases to identify high-probability candidates before experimental evaluation.
Submit your inquiry to request a custom solution.
References
If you have any questions or encounter issues on this page, please don't hesitate to reach out. Our support team is ready to assist you.